The creation of cell-based disease and research models can inform drug development programs through mechanism elucidation, isogenic disease models, loss or gain-of-function studies and more. Because of the imprecision and variable transgene expression associated with random integration, many times development of models with these technologies is delayed or comes to a complete halt. On the other hand, popular single-guided RNA methods such as CRISPR-Cas9 have off-target risk and complicated licensing hurdles. Hera BioLabs has set out to develop an efficient strategy for site-specific transgene integration and editing in cell lines.
Targeted Editing And Improved Transgene Consistencies With Cas-CLOVER
If you’re wanting to optimize your research models, Hera’s Cas-CLOVER platform enables efficient insertion for gene expression and endogenous gene editing at any target loci in mammalian cells. By working with Hera’s team you will quickly be able to produce more effective research models tailored to advance your programs. In this article, we would like to focus on targeting the Hipp11 (H11) gene, as it is well characterized as a “safe harbor” locus for gene knock-in within CHO-S cells.
Before getting into the details, let’s review the identification of the H11 locus and why it’s considered an effective integration site. Genomic loci that are considered “safe harbor” are sites that are known to be very effective for transgenesis, and result in high and consistent expression. Knock-ins at safe harbor sites do not negatively affect the characteristics of the cell.
Located between the Drg1 and Eif4enif1 genes, the Hipp11 locus offers tremendous potential for providing stable gene knock-in and high expression.[1, 2] The robustness of H11 has been clearly demonstrated in multiple animal models, including mice, pigs, human embryonic stem (hES) cells, and induced pluripotent stem (iPS) cells. [1–3]
Hera set out to establish Cas-CLOVER targeted knock-in reagents and protocols at the Hipp11 safe harbor locus. Our proof-of-concept (POC) strategy is shown below and requires the knock-in of a 4kb piece of DNA.
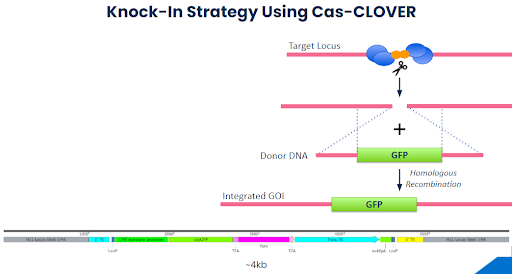
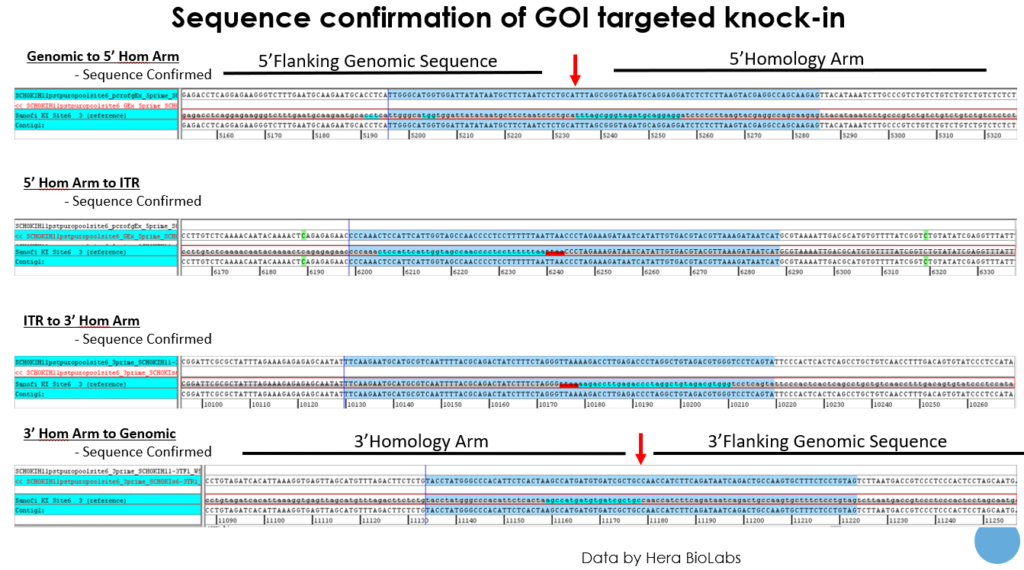
Using the Cas-CLOVER system, our results have shown that one can achieve remarkably high targeted knock-in efficiency in as little as two weeks. The knock-in efficiency of our 4kb transgene was estimated to be 80-90%, following a short 10-17 day cell culture and selection process. The targeted integration at the H11 safe harbor site was clearly confirmed by sequencing.
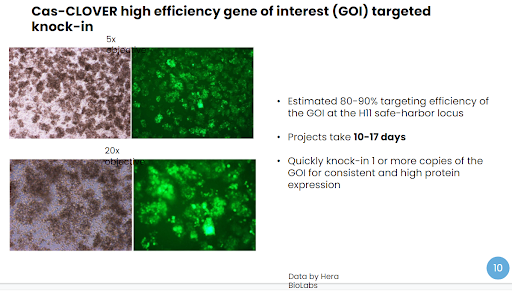
In comparison, random insertion yielded 0.68% GFP-positive cells in this study of CHO-S cells, and 1.32% GFP-positive cells in HEK293T cells. Targeted integration by using traditional CRISPR/Cas9 is an option, as one study showed an indel frequency of 43% at H11.[4] Alternatively, the Cas-CLOVER platform takes optimizing your research models to the next level. Cas-CLOVER has already demonstrated more accuracy than traditional CRISPR/Cas9, with 0% off-target mutagenesis. In contrast, CRISPR/Cas9 has shown upwards of 10% unwanted cutting in off-target analysis with CHO cells. This level of off-target mutagenesis impacts research cell lines negatively by creating additional, unintentional and unknown phenotypes., By utilizing Cas-CLOVER for effective and precise editing, together we can optimize the research models used in your discovery programs.
Cas-CLOVER Cell Editing
Interested in Hera’s Cas-CLOVER editing services and technology licensing? The Cas-CLOVER targeted nuclease system is one of the most efficient platforms for cell-line modification. Speak with Hera to learn more about our services.
Learn more about Cas-CLOVER system by contacting us today.
