As cell line development bioprocessing turns from traditional random plasmid integration to targeted knock-ins for maximizing yield and stability, it is important to choose the right gene editing tools. The RNA-guided Cas9 nuclease from Streptococcus pyogenes (SpCas9), which is the most commonly used enzyme in genome-editing applications, can yield significant levels of off-target activity, with reported off-target cutting as high as 13%. Multiplex editing with SpCas9 for cell line engineering can also lead to high rates of translocations, up to 4% for individual translocations or rearrangements and up to 5% cumulatively.1
A recent study published in Molecular Therapy Nucleic Acids saw the authors at Poseida Therapeutics deploy the dual RNA-guided dimeric endonuclease Cas-CLOVER, as an alternative to CRISPR/Cas9. Hera and Poseida both use Cas-CLOVER technology in different fields of use. Let’s take a closer look at the targeted knock-in data in this study with Cas-CLOVER.
Comparison of knockout and knock-in success in iPSCs between Cas-CLOVER and Cas9
In this study, the team determined HDR (homology-directed repair)-mediated knock-in potential using Cas-CLOVER by examining reporter cassette delivery at several loci (GAPDH, B2M, and HBB) in induced Pluripotent Stem Cells (iPSCs).
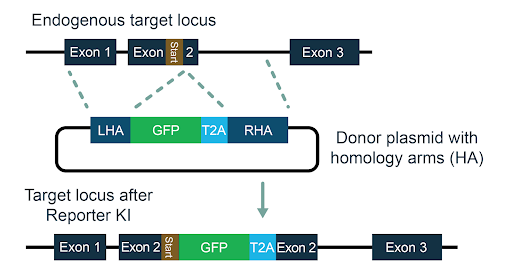
Figure 1: Schematic showing targeting strategy.
1 Cas-CLOVER is a novel high-fidelity nuclease for safe and robust generation of TSCM-enriched allogeneic CAR-T cells. Madison et al.Molecular Therapy – Nucleic Acids. August 8, 2022.
At most loci, Cas-CLOVER’s indel – as well as knock-in – efficiency is equal to or greater than that of Cas9, as demonstrated in the below example for B2M.
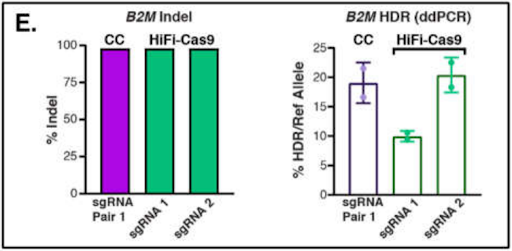
Figure 2: Quantification of indel (left) and HDR rates (right) at the B2M locus.
At HBB, knockin efficiency was higher using Cas-CLOVER, but the difference was probably not due to differential efficiency of target cleavage, as gauged by Cas-CLOVER’s lower indel generation (see image below). Rather, the higher efficiency was likely a result of the larger deletions, with overhangs, that are characteristic of Cas-CLOVER. Cas9 indels generally result in small 1-3 bp blunt-ended cuts.
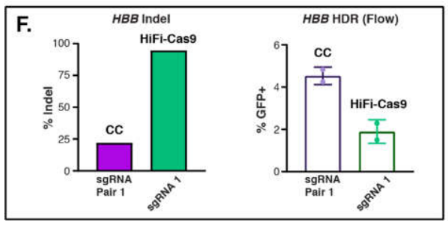
Figure 2: Quantification of indel (left) and HDR rates (right) at the HBB locus. Flow for HDR measurement at the HBB locus was performed 14 days post-nucleofection to ensure episomal GFP expression from the donor plasmid had decreased below the detection limit.
After Cas-CLOVER editing, potential off-target sites were queried for the presence of indel mutations. Indel frequency at off-target sites did not occur at a statistically significant level above background. The team concluded that, within primary T cells from multiple donors, the Cas-CLOVER platform performs as well as any other targeted nuclease, but with much lower off-target activity. Notably, Cas-CLOVER yields efficient multiplexed gene editing.
Targeted knock-in capability is becoming essential for cell line development
Induced pluripotent stem cells (iPSCs) are being used more and more in therapeutic bioprocessing and in synthetic biology. Moreover, the level of targeting that the Poseida team was able to achieve with Cas-CLOVER is relevant for any cell line development professional – including those engineering custom cells for preclinical research purposes.
Learn more about Hera’s Cas-CLOVER and piggyBac technology
To learn more about Cas-CLOVER, piggyBac, and how Hera can help accelerate your gene editing research with our toolkit, click here. We have everything for your gene editing needs, including flexible front or back-loaded service terms and licensing options. Access our technology as quickly as possible by shopping online today.
References
- Cas-CLOVER is a novel high-fidelity nuclease for safe and robust generation of TSCM-enriched allogeneic CAR-T cells. Madison et al.Molecular Therapy – Nucleic Acids. August 8, 2022.
