Tal-CLOVER
Hera utilizes Cas-CLOVER as our preferred method for gene editing technology for many reasons, including that Cas-CLOVER is simple to design and build and has extremely high efficiencies and target fidelity. However, not all cell types or gene targets are amenable to gRNA-based targeting (either with CRISPR or Cas-CLOVER).
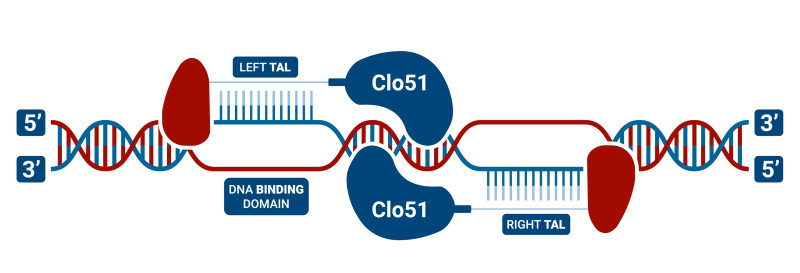
TAL-CLOVER is a TAL DNA binding domain with our unique Clo51 dimeric restriction endonuclease (rather than a Fok1). These are highly flexible for design since it only requires a “T” anchor and this results in more plentiful anchor binding sites especially in TA-rich regions of the genome. Another advantage is that TAL-CLOVER, like Cas-CLOVER, requires enzyme dimerization and thus has high fidelity.
The main downside is that the TALs in TAL-CLOVER are large, generally hard to build, and labor-intensive. Hera uses our proprietary YESHI process to rapidly construct TALENs in as few as 10 days. Now, we are offering this version of our gene editing technology as an option to our clients when other technologies aren’t a viable option.
What is a TALEN?
TALEN (Transcription Activator-Like Effector Nucleases) structure is largely similar to zinc-finger nucleases (ZFNs) in that they are composed of heterodimers of a DNA binding domain and the nuclease of the dimeric restriction endonuclease. In our case, the heterodimer is the Clo51 endonuclease, while other groups use the Fok1 version. However, instead of using a zinc-finger protein for DNA binding, they use Transcription Activator-Like Effector (TALEs), which were originally identified in plant pathogenic bacteria. TALEN technology consists of tandem repeats of 33-35 amino acids, of which each binds to a single DNA base-pair. TALEN genome editing has become more prevalent than the use of ZFNs, as they overcome a number of obstacles such as decreased cytotoxicity and the ability to target nearly any DNA sequence. Some studies also indicate that they have reduced off-target mutation rates and mediate higher homology-directed repair compared to other site-specific nucleases, including CRISPR/Cas9. There have been a number of studies that have successfully utilized TALEN technology in hPSCs. Like ZFNs, this technology has also been used in combination with the piggyBac system to facilitate the seamless removal of the donor cassette. TALENs have also been used in combination with single-stranded Oligodeoxynucleotides (ssODNs) to create point mutations in the genome. This technology continues to be a popular choice for gene editing because unlike ZFNs or CRISPR/Cas9, TALENs can bind to nearly any DNA sequence. This, therefore, provides a clear advantage for nuclease-design strategies.
TALENs vs. CRISPR/Cas9
TALENs ( including our TAL-CLOVER) and CRISPR-Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats-associated protein 9) are both powerful gene-editing tools that have revolutionized genetic research and therapeutics. However, they do have different properties, strengths, and weaknesses.
Ease of Design and Use: CRISPR-Cas9 is generally easier to design and use. The system uses a small piece of RNA to guide the Cas9 nuclease to the target DNA sequence, making it relatively straightforward to design new guide RNAs for different targets. In contrast, TALENs require the design and construction of new proteins for each target, which can be more complex and time-consuming.
Specificity and Off-Target Effects: TALENs are often more specific than CRISPR-Cas9 and have fewer off-target effects, which are unintended modifications of DNA sequences other than the target. This is particularly important in therapeutic applications, where off-target effects could have detrimental consequences.
Efficiency: Both TALENs and CRISPR-Cas9 have similar efficiencies in introducing breaks in the DNA, but efficiency can vary depending on the specific target sequence and the cell type.
Cellular Delivery: Both systems can be delivered to cells using similar methods, such as plasmid transfection or viral delivery. However, the larger size of TALENs can sometimes be a limitation for certain types of delivery systems.
Multiplexing: CRISPR-Cas9 is generally better suited for multiplexing, or targeting multiple genes at once, as it is relatively easy to design multiple guide RNAs. In contrast, multiplexing with TALENs is more complex as it requires the design and construction of multiple proteins.
In summary, the choice between TALENs and CRISPR-Cas9 often depends on the specifics of the project at hand. For example, if high specificity and fewer off-target effects are critical, TALENs might be the better choice. However, for ease of use, flexibility, and multiplexing, CRISPR-Cas9 often comes out on top. It’s also worth noting that these two technologies are not mutually exclusive and can often be used complementary in a research setting.
Singh et al. (2015) Gene Editing in Human Pluripotent Stem Cells: Choosing the Correct Path. J Stem Cell Regen Biol.
Cas-CLOVER TM
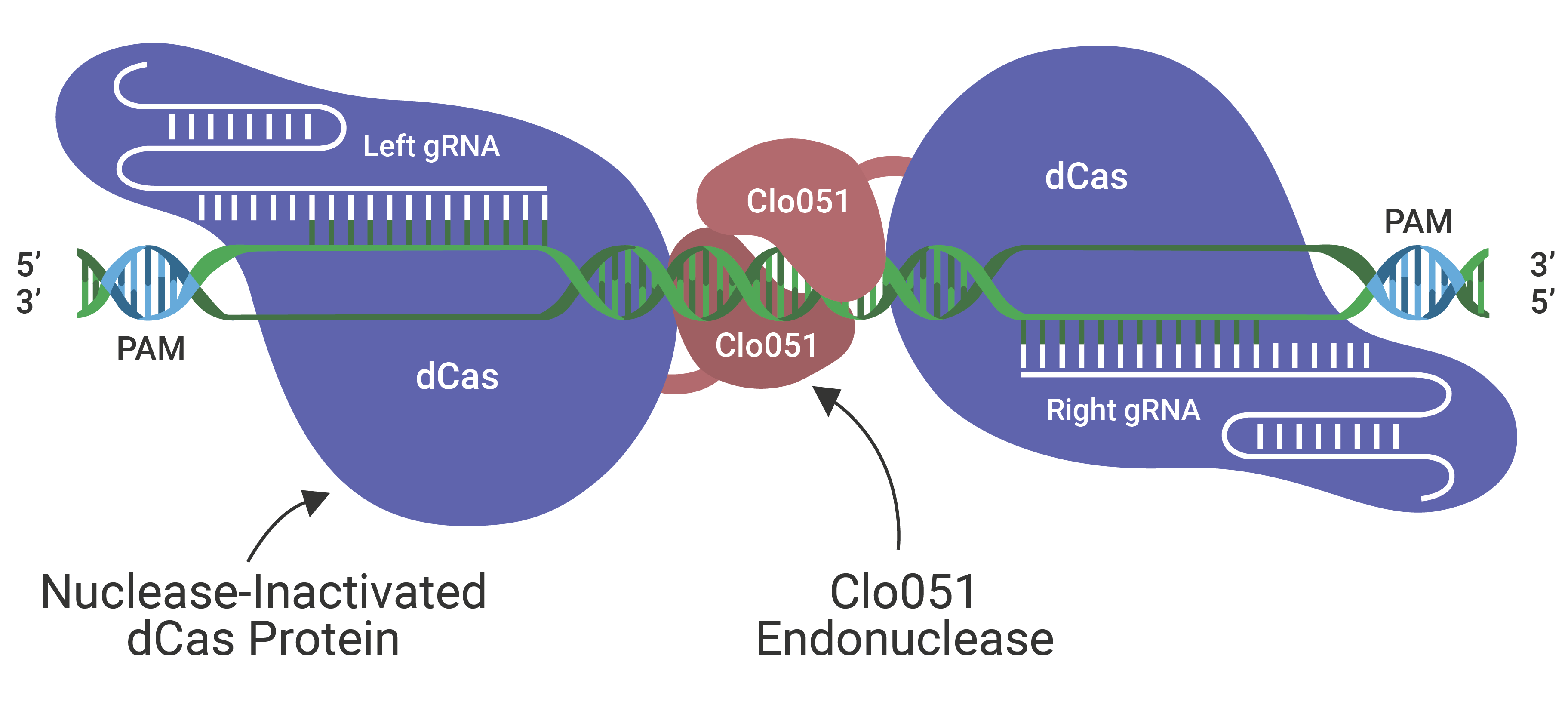
The Cas-CLOVER Site Specific Nuclease Technology is our highly specific dimeric nuclease technology for targeted gene modifications. With this technology, we engineered our GS (glutamine synthase) knockout suspension CHO cell line.
Cas-CLOVER has been optimized for high efficiency on-target nuclease activity with low to zero off target mutagenesis. The increased specificity is the result of using a pair of gRNAs and the obligate dimer CLO-51 for the enzymatic activity.
We have freedom to operate using our proprietary patented Cas-CLOVER technology, allowing Hera to provide custom cell line development services with flexible and clear commercial terms.
